Research Projects
Mining Time Evolving Networks
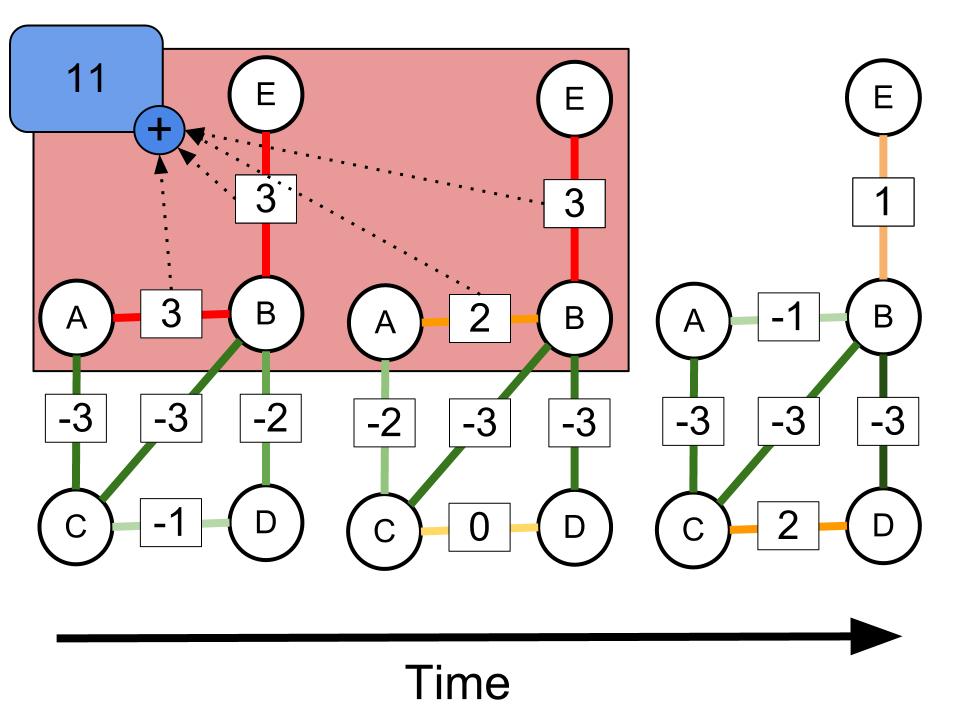
Time evolving networks arise in multiple domains. They characterize traffic variations in transportation networks, information flow in communication networks, variation of trade rates in a network of trading agents, or phases of pathway switching in gene interaction networks. One model of a time-varying network is based on fixed network structure (edges and nodes) and a time-varying characteristic of the nodes and edges. Within this setting, significant or unusual patterns are defined as temporal network regions whose behavior deviate from expectation. There are different ways of defining such aggregate patterns of interest based on whether graph region corresponds to a fixed subgraph or one that varies across time (and constraints on variations). Furthermore, of interest is the discovery of higher-level patterns such as splitting, merging, and shifting, and languages to define such patterns.
Nanomaterial design and discovery
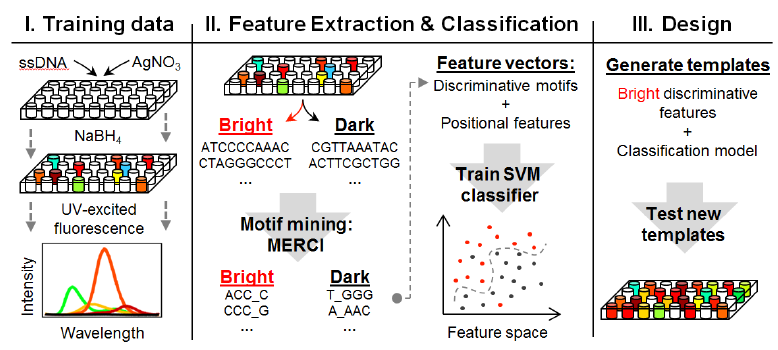
DNA-stabilized silver clusters (Ag-DNAs) are novel fluorophores that are finding numerous applications in nanophotonics, chemical sensing, and bioimaging. The fluorescence colors of Ag-DNAs can be tuned from blue-green into the near-infrared by selecting the sequence of the single-stranded DNA that templates the cluster. Using a training set of DNA template strands and the fluorescence spectrum associated with each strand, we mine discriminative multi-base DNA motifs that correlate with fluorescent cluster brightness. Furthermore, using such motifs to parameterize DNA templates, we develop a machine learning-based tool to design novel DNA templates that stabilize brightly fluorescent Ag-DNAs.
Gene Networks
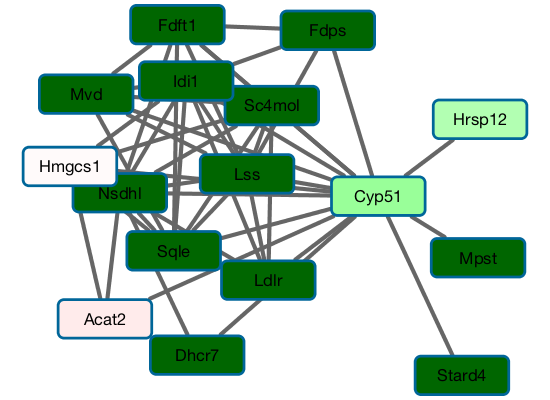
A gene network models the interactions between genes in the cell. Beyond their topology, such networks feature local gene states corresponding to expression levels under a given experimental condition and global states corresponding to a specific phenotype. In this setting we are interested in novel algorithms for global state prediction based on subnetwork states as well as unsupervised detection of gene subnetworks that deviate in their local states from the rest of the network.
Previously we also considered the problem of gene function prediction. We proposed a two-phase approach for the problem - feature extraction from a partially labeled network coupled with classification. The approach is robust to the network structure and dominates existing alternatives.
Social Media Analysis and Optimization
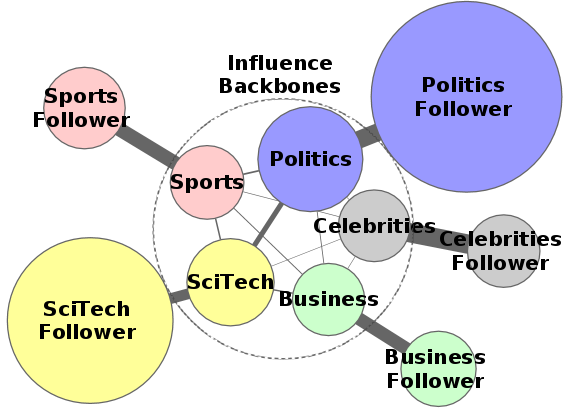
Social media provides a novel paradigm of disseminating information by shifting the authority from traditional media outlets to practically anyone with an Internet connection. This new paradigm poses a multitude of interesting scientific questions, but the grand outcome is that we finally have socio-computational systems that evolve and behave on their own as a result of the participation of millions of users. The goal of this project is to investigate how people and their individual behavior contribute to the global network behavior that emerges. Some important problems include deriving a per-user fingerprint (genotype) of interaction with information on various topics, predictive models for user opinion, detection of issue-specific activity bursts and others. Other questions in this project are related to network design: How to optimize desirable network properties such as fast information propagation and identifying subsets of collectively central nodes?
Brain Networks
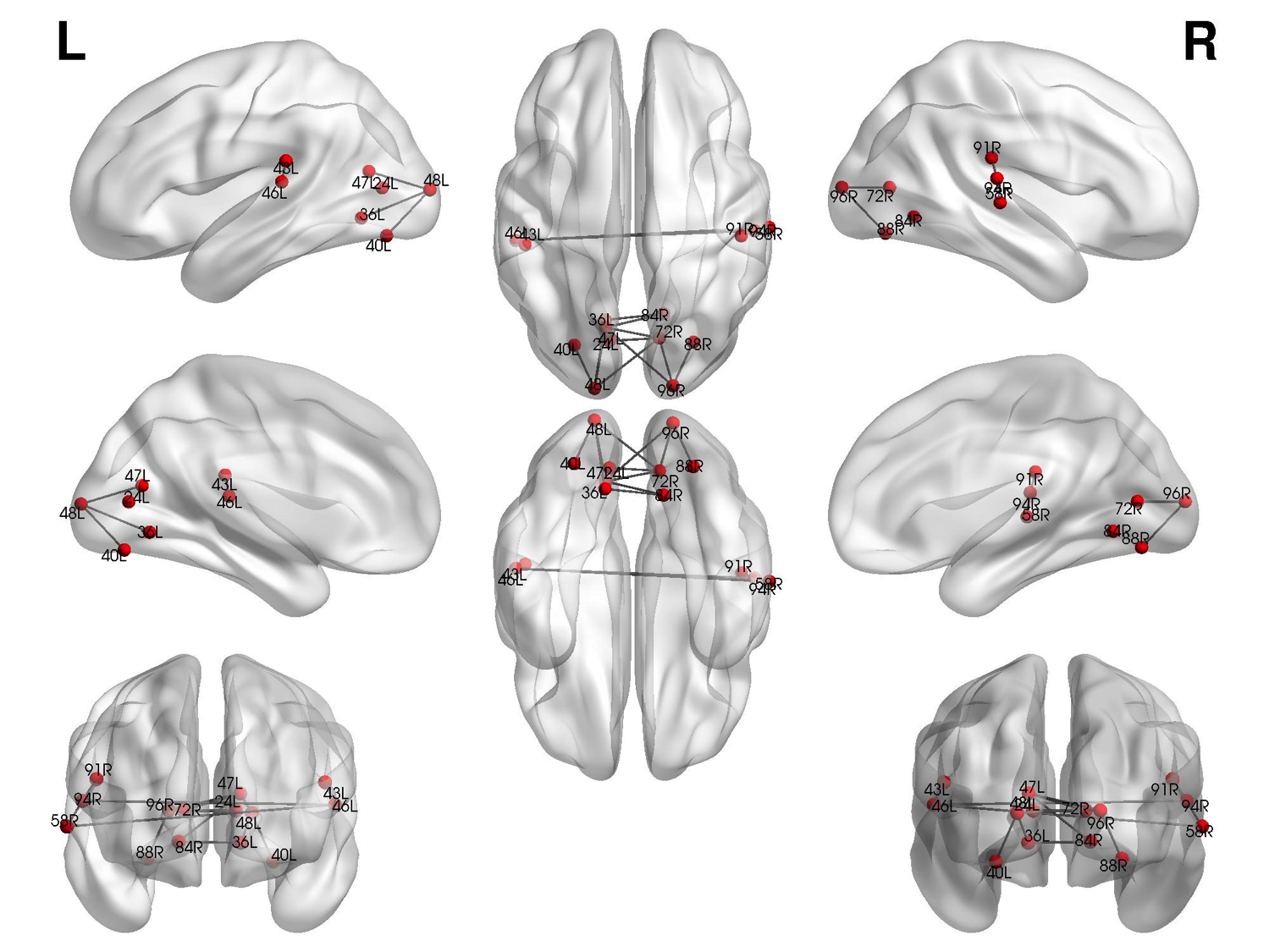
The goal of this project is to identify subnetworks in the brain that are predictive of cognitive behavior such as sensori-motor learning as well as disease states. Functional brain networks are created from fMRI measurements during a cognitive activity or for different patients. The functional edges among brain regions are characterized by the level of their coherence (co-activation). The task at hand is then: How to identify significant subnetworks whose coherence is predictive of the performance in cognitive task or existence of a neurological disorder.
